Project: Life-Chip – Modular Biopsy-on-Chip Development
Life on a Chip develops a modular, customizable microfluidic biopsy-on-chip platform and a test station that allows users to independently verify device reliability before use.
The project aims to strengthen personalized medicine by enabling reproducible and scalable research on patient-derived tissue.
We thankfully acknowledge that this project is co-financed by the European Union – Just Transition Fund (JTF) and supported by SNN under the VIA Groningen 2025–2026 program.
Project duration: September 2025 – March 2027.
🔹 Funded by the European Union – Just Transition Fund (JTF)
🔹 Supported by SNN


Showcases (July 2025)
Microfluidic Biopsy Holder: Enabling Long-Term Biopsy Studies
By Dr. Axel Hochstetter — Life on a Chip & Zosia Komar — Erasmus MC
Introduction
The Microfluidic Biopsy Holder enables long-term biopsy viability through continuous perfusion, allowing for real-time observation and comprehensive polyomic analysis (metabolomics and proteomics). Designed for ease of use and precision, it fits standard microscopy holders and can be adapted for various biopsy shapes and sizes.

Application samples
- Cancer Research: Study tumor biopsies over extended periods.
- Polyomics Studies: Collect biochemical data while keeping the sample perfused.
- Drug Testing: Observe biopsy reactions to drugs.
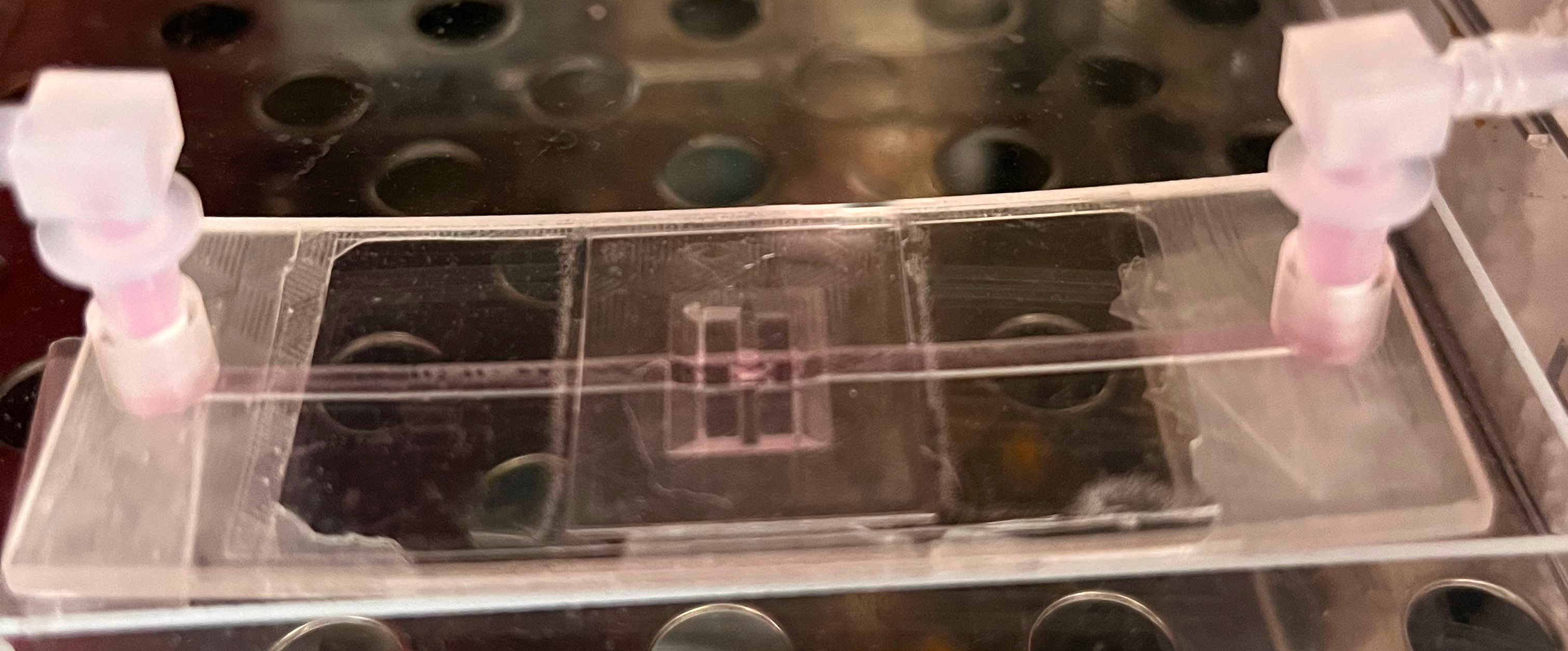
Features
- Fits your biospies: (e.g. 5 mm long, 1 mm diameter for biopsies of skin or throat cancer). Different size? Just contact us!
- Easy sealing, re-opening, and re-sealing: Allowing you to retrieve and e.g. (re-)stain the biopsy, and re-use the holder.
- High optical transparency: compatible for fluorescence, confocal and Raman microscopy.
- Contability with microscopy stages (75 x 25 mm²) and microtiter plate readers (127.76 × 85.48 mm²).
- Easy connectivity: individual connectors are built in (e.g. for Luer-Lock, tubes, etc…).
- Biocompatible material: plant-based acrylate, for high-end applications, a parylene coating is possible to completely shield your sample.
- Disinfected Packaging: Ready fror use in clean environments. Autoclavable material options available.
1. Cell sorting in microfluidics (via DLD)
Introduction
Microfluidic devices have been used to separate a variety of target cells and particles from samples like blood, urine or cerebral spinal fluid.
One approach to this separation uses an impressive, passive microfluidic technique called deterministic lateral displacement (DLD for short). The beauty lies in its simplicity: Once you have built your array, the sorting is basically a by-product of the sample flowing through the device. Your perfectly tailored microfluidic environment can run continuously, separating your desired targets (e.g. circulating tumour cells, parasites, marker DNA, exosomes, etc.…) from anything else in your sample.
So, why is it not widely used?
Well, it is not that simple (of course!)
Naive model
First of all, even the simplified „naive model“ (as Timm Krueger and I love to call it) takes a bit time to think through there:
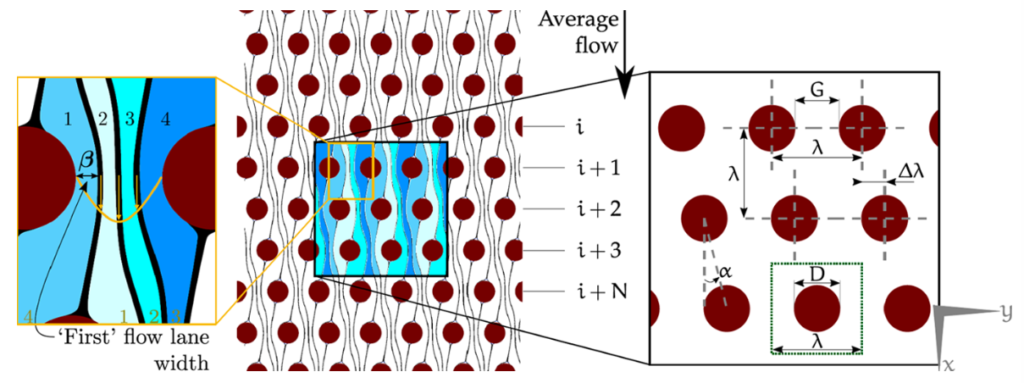
So, basically, your sample flows through an array, that is tilted in respect to the main flow. The smaller the tilt, the more lanes your flow is divided into. The more lanes, the smaller is each lane – especially around a post of the array. Now, if your particle is significantly bigger than the lane it travels in, it gets pushed into another layer and cannot follow the flow anymore. In this picture, it gets bumped to the right (spoiler: look at the last picture of this post. There a parasite gets bumped downward, while blood cells can continue to flow along horizontally).
Plethora of parameters
Secondly, there is a number of parameters that influence the DLD performance:
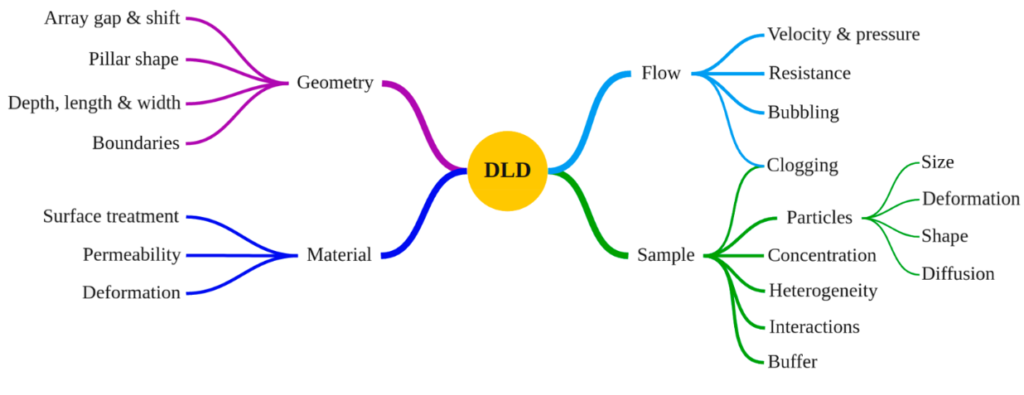
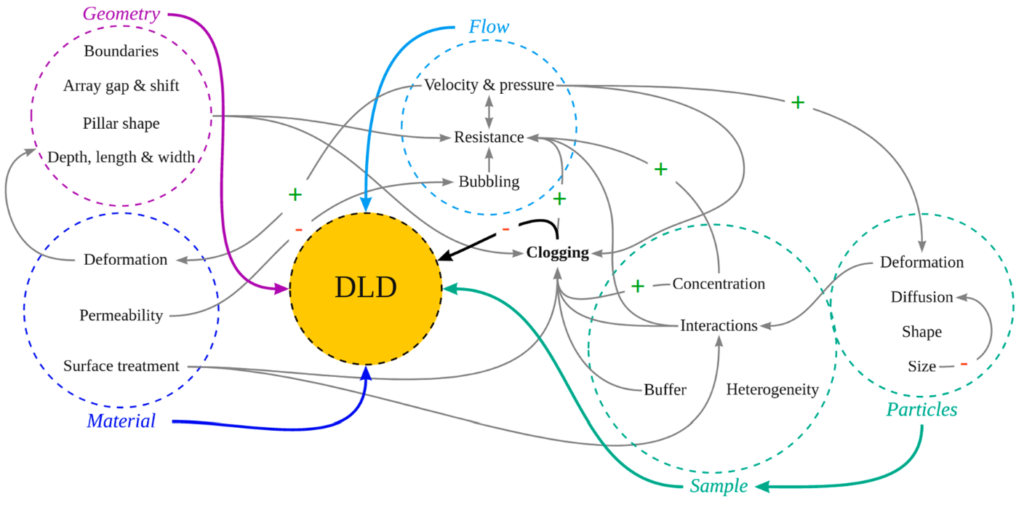
BUT: Although it is complicated, it is not impossible:
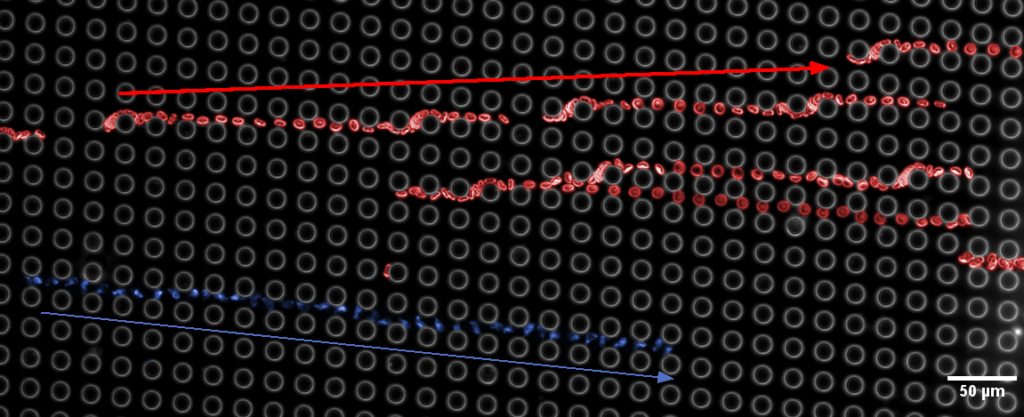
If this sounds intriguing, and you wonder whether DLD is right for you, contact us!
We will get you sorted!

2. Microfluidics for drug discovery and 3R
Microfluidics has many applications. One of these is surprisingly under-used – despite the potential to save billions of Euros and years of research time, and to save thousands of (animal) lives: Organs-on-Chip (OoC)
OoC are miniaturized models of (human) organs. They are kept in a highly controllable, microfluidic environment.
Since they have the full functionality of organs, they can be used in drug development (metabolizing drugs, screening for cytotoxic drugs and metabolites). Simultaneously, their massively smaller size and independence from other organs makes them cheaper to grow and culture, and reduces the amount of nutrients and drugs needed.
For pharmaceutical companies, OoC is the clever step to take:
Out of 10 potential drug /target molecules that are successful in cell culture and mouse model test, 9 fail in human testing, having cost around 2.5 Billion $ and 10-12 years. Why do 90% fail? Because of the differences between mice and men.
Here, human Organs-on-Chip can help! With a setup running in parallel or even before the animal models, these 9 out of 10 duds can be screened out, saving billions of Euros or Dollars and years of time, allowing us to focus on the 1 candidate, that will also actually work in humans.
Balancing these billions of Euros with the initial costs to use OoC (which is between tens of thousands to a few millions) it is surprising that so few have done it!
But how would an Organ-o-a-Chip look like?
Let’s illustrate that with a potential drug that needs to
- A) cross the blood-brain barrier (or any other kind of tissue: lung tissue, epidermis…) and
- B) affect a certain kind of pathogen there
This can be done in the following sample device:
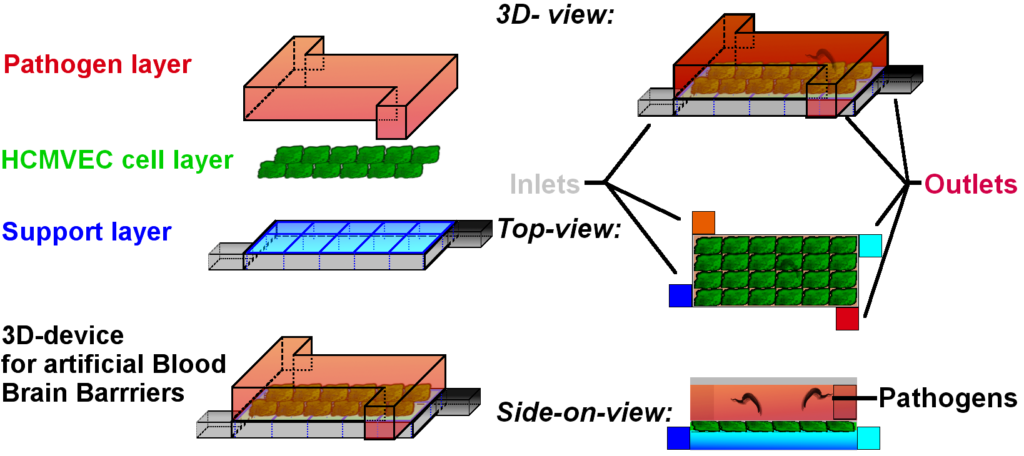
This device is made of two halves (top and bottom) separated by the tissue we want to culture inside. The cells are growing on a support layer, that can also supply the cells with medium and wash away metabolites. The top layer however would be, where the evil pathogens abide.
The cells are grown in a confluent blood-brain barrier* and keep the pathogens at bay.
Now, we can inject the drug candidates into the support layer, and see
- A) whether the pathogens will be eliminated
- B) how fast this happens
- C) whether the cells themselves take any damage.
*: It should be noted that this is a simplified scheme. Making a functional blood-brain barrier already requires co-culture of more than one cell type. The details on how to grow these cells, and how to feed the inlets and take away the waste would be way too much for here. But if you are interested, contact us at info@life-chip.com, and we will get you started on the process. Promised!
This is, as mentioned, still a rather simple and straightforward example for an Organ-on-a-Chip.
Of course, one can have several organs in a row, to study the interplay of drugs and metabolites or infections. There are several companies currently working with two or three interconnected organs on one chip. One can even grow an entire „Human-on-a-Chip“ to see the full metabolism (e.g., lung – gut – liver – heart – bone marrow – kidney, see Figure 2) of a drug.
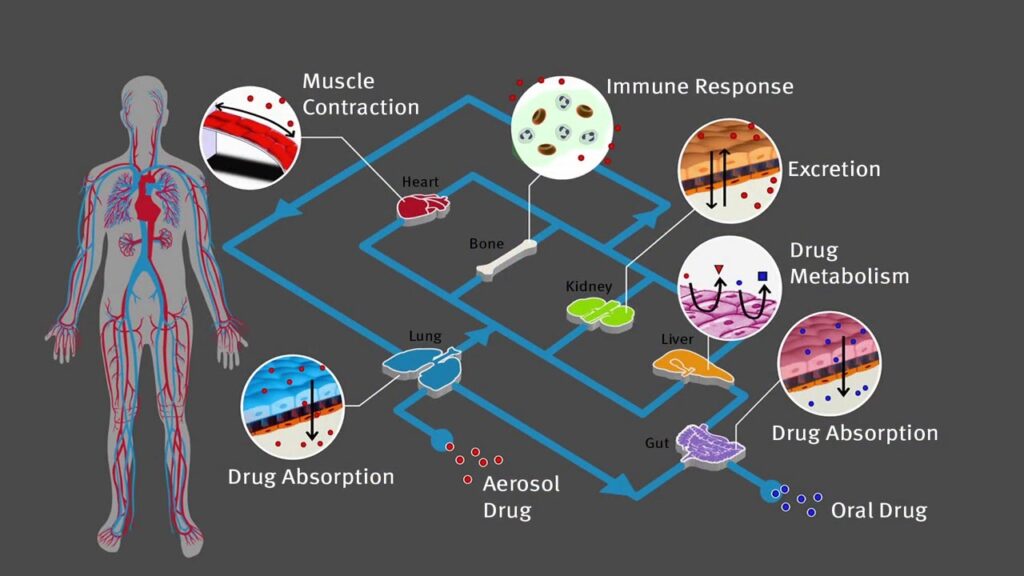
But for most applications, that is going too far, and a simpler approach can already yield very good results. This means, with even just 2 or 3 OoC you can screen off 7-9 of the 10 candidates that work in mice, but not in humans. Of course, there are challenges: co-culturing of different cell lines with their own nutrition and growth requirements, handling of several fluid streams, etc…
But in general, here are the Pros and Cons one should consider:
| Pros: | Cons: |
| Potential to screen out ineffective candidates before they reach animal or human trials | Initial investment necessary |
| No animals (license) needed | In-depth knowledge needed to set up the lab equipment and train staff (this can be outsourced to „Life on a Chip“ though; info@life-chip.com ) |
| Possibility to use actual human cell lines for human targeted products | |
| Smaller sample volumes needed | |
| Faster results | |
| High concentration control | |
| Saves money and time | |
| Possibility to assess different cell lines (human / mouse/ rat/ rabbit) in parallel |
Some of the often overlooked features of Organs on a Chip are
- it is possible to use several organs in sequence
- it is possible to grow targeted inflamed/damaged/infected organs – and study the polyomics of that
- it is compliant with 3R and thus makes your project eligible for funding
- it is an additional trial phase that can screen out ineffective drug candidates prior to animal or human trials.

